|
Last update 7 March 2016.
Updated with another example Febr 17 2013.
|
 |
While analyzing the data in the file CO20050101-LCE-AjC.txt at the French
CNRS database
I observed a number of quite high TTest correlation values which were not considered as correct datings.
(If the file is not there any longer, try to search for that filename!
|
|
 |
|
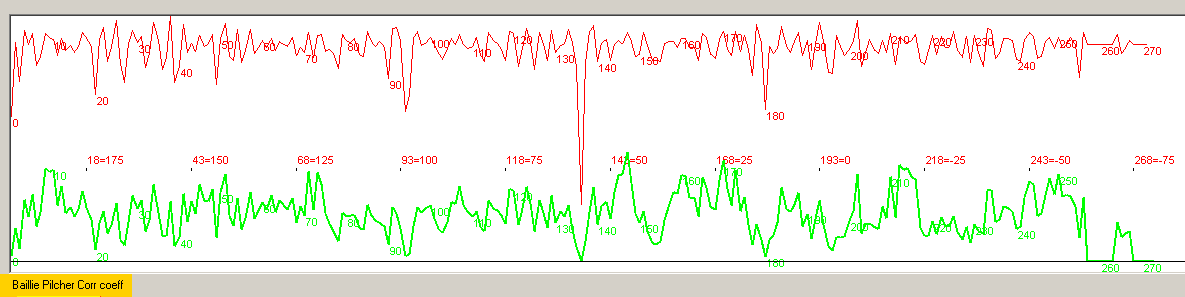
A look at the ring width data of some of the files revealed that the lab uses the convention to
register very narrow rings with the value "1" as shown above. With these values the surrounding
rings will then have values which are between 50-200 times as wide as that narrow ring. (Missing
rings - are marked with a ",")
|
|
 |
|
The wide span in values will make the Baillie/Pilcher normalized curve (the red curve at the top above) dominated by the narrow ring.
|
|
 |
|
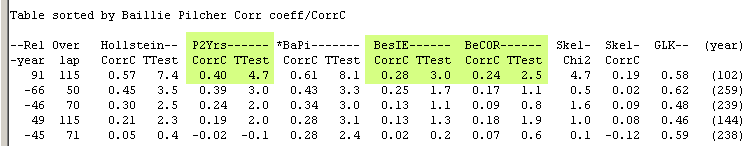
When two curves, which both have such a narrow ring, are crosscorrelated with either the Baillie/Pilcher normalization
method (as above) or the Hollstein method, these narrow rings will "attract each other", i.e. give a match at the wrong position.
|
|
 |
|
Note that the other normalization methods are not fooled by the
existence of these extreme narrow rings!
|
|
 |
Let me show still one more example:
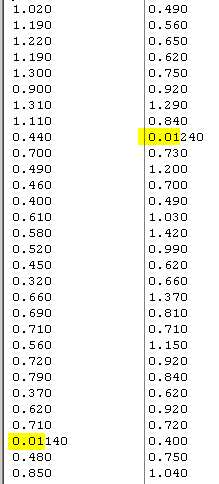
Above is ring width data collected from two Scots pines in 1995.
The data has been somewhat manipulated, by changing one of the
narrow rings from 0.140 mm into 0.01140 mm and another narrow ring from 0.240 mm into 0.01240 mm
|
|
 |
|
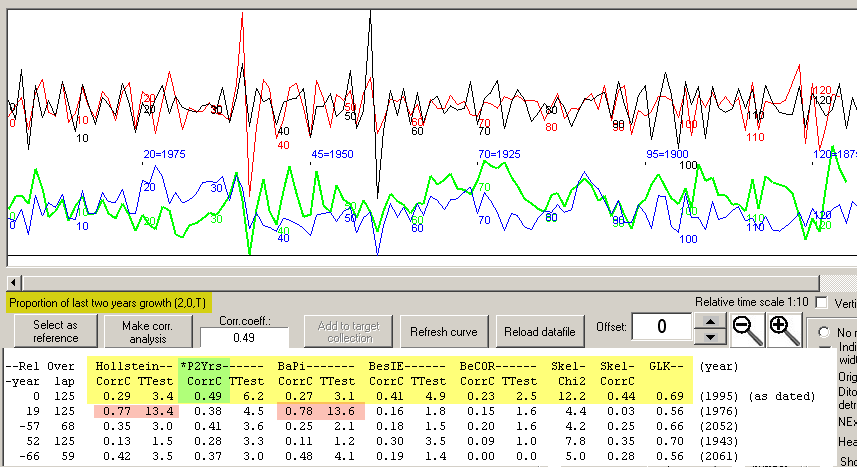
When these two manipulated ring width curves are crossdated towards each other with the
Prop2Yrs (Proportion of two last years growth) method, the correct dating will be "top ranked"
as shown above. Though the Baillie/Pilcher and Hollstein methods will get fooled by those
two very narrow rings!
13 April 2009, Lars-Åke Larsson
|
|
|
|
Here is an example which we found within a 7600 years long pine collection from Finland. While
rebuilding the whole collection of some 1200 selected members of enough length and good matching we found
one single sample which at the first glance looked incorrectly dated. I.e. during the rebuilding (crossdating) process
CDendro proposed quite another dating than that used for the sample within the collection.
|
|
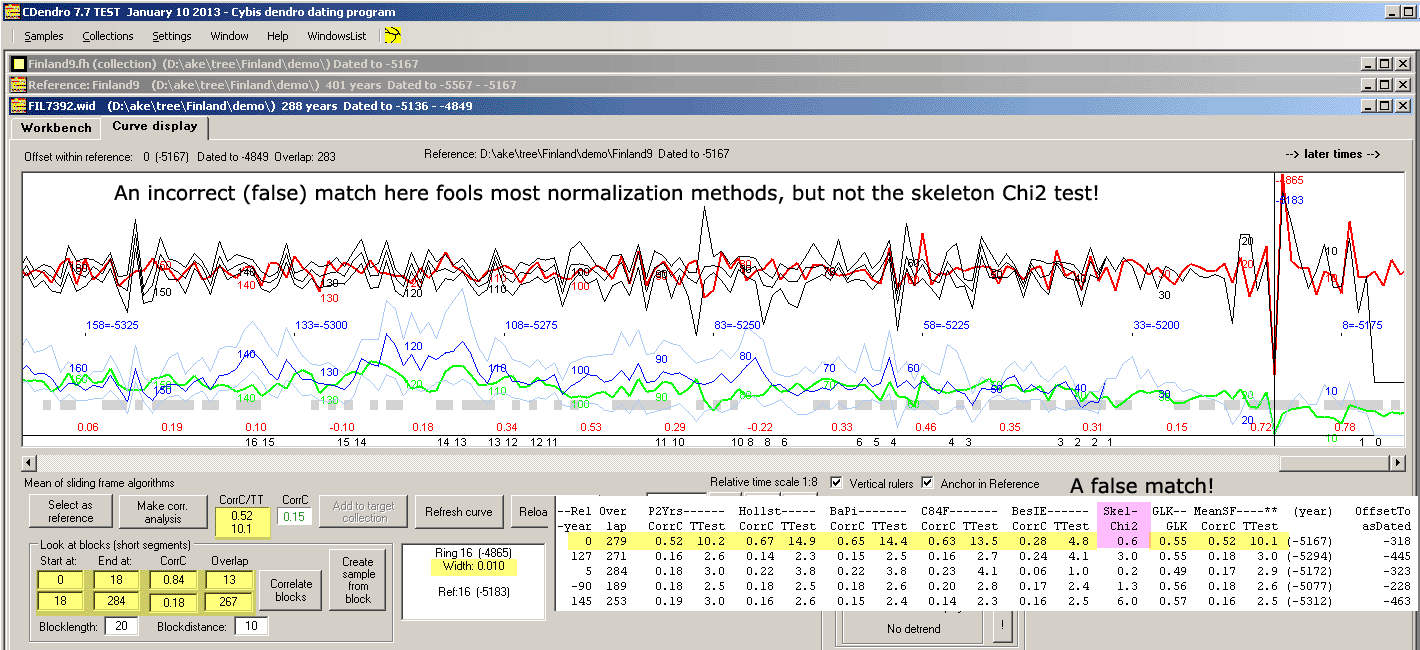 |
|
The picture shows the unexpected match between a subcollection (as a reference) and a single sample. The mean correlation values from several
normalization methods says 0.52/T=10.1 with 279 years overlap. I.e. the T-value looks suffieciently high to make the match credible!
But the skeleton Chi2 value is very low, only 0.6 and the curves do not look as really matching each other!
Actually a 267 years long overlapping section has a correlation coefficient of only 0.18/T=3.0
What has happened is that the reference and the sample both have a very narrow ring.
For some normalization methods, such narrow rings "attract" each other during the correlation based selection process!
It should be mentioned that Cofecha running with standard settings, does not get that fooled
and reports a correlation coefficient of only 0.28 corresponding to a T-value of 4.9 and also
reports on many segments with possible problems.
|
|
There are two obvious methods to avoid the problem above:
- Use a normalization method which is not that sensitive to narrow rings.
- When selecting a best sample for good crossdating, require not only a high T-value, but also a "good enough" Skeleton Chi2 value.
Both methods are already implemented in the current TEST version of CDendro 7.7 (January 2013):
- A new normalization method: "P2YrsL Proportion of last two years growth LIMITED" where all peaks
within the normalized curve are by default cut at 2.6 times the standard deviation for the curve.
- The tool "Add best members to target collection" now has a textbox for requiring a minimum Skeleton Chi2 value before a sample is presented as matching the reference.
|
|
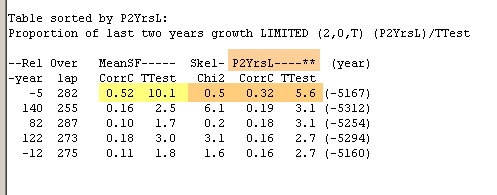
|
The "peak cutting version", P2YrsL, now shows that false match with CorrCoff=0.32 to be compared to Cofecha's lower - and thus better value - of 0.28.
|
|
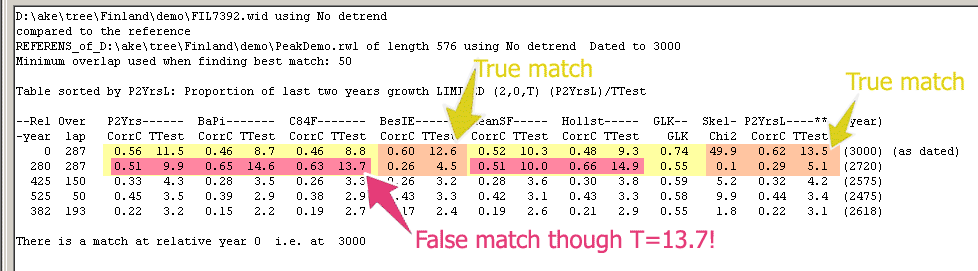 |
|
Finally, I have mounted a part of the reference section used above together with a segment containing the correct matching point and given them a fictitious dating.
A false and a correct match are then shown in the table above where some normalization methods give a considerably higher T-value for the false match than for the correct one.
For your testing, here is a .rwl file containing both the reference (memberId=AsRef) and the sample (memberId="sample"):
Test data for download: PeakDemo.rwl.
Many thanks to Mauri Timonen for supplying me with this interesting data!
Lars-Åke Larsson
|
|









